Synthetic Turbulence Inflow Generator
Synthetic Turbulence Inflow Generator (STG)
Contents
Overview
The Synthetic Turbulence Inflow generator is a method to produce an inflow boundary condition that results in turbulent flow in a short downstream distance. This method is desired to be more computationally efficient than resolving a flow through from transition. This page will cover the workflow to produce the necessary auxiliary files as well as included specifications for the solver.inp file for running a simulation with the STG boundary condition. There are three auxiliary files that are needed for the method in the n-procs_case directory at runtime:
- STGInflow.dat
- STGSpectra.dat
- STGRand.dat
In addition, there are scripts to determine the convergence of the 1st and 2nd order mean profiles that PHASTA will simulate BEFORE PHASTA simulates it. All of the mentioned tasks are performed with MATLAB scripts located in the STGInit subdirectory of the file structure contained in the PHASTA code-tree from github.
STGInflow.dat
This is a file that will contain the mean velocity profiles, mean cross correlation targets, and any additional turbulence modeled scalars.
STGSpectra.dat
This file will contain the N spectral modes for each node that has the STG the boundary condition.
STGRand.dat
This file will contain random numbers for the N spectral modes. This is not a spatially varying set of numbers as the spectra is.
README
Within STGInit subdirectory, there is a README file that contains a list of the most important routines, their inputs, their outputs files, and their outputs that occur in the command line. These scripts are listed in chronological order of the workflow. In addition it lists the helper functions that are also needed but do not require intimate knowledge for the ordinary user. This README is below:
1 README for the matlab scripts for STGRand generation 2 3 4 Below is a table that summarizes the inputs and outputs of the main matlab scripts developed. Further explaination is provided in the 5 header of each file. The order of the script in the table below corresponds to their order in the workflow, I.E. choose your random numbers, 6 find the long term effects of those choices, plot them, plot the time convergance wrt the bias, and re-calc bias measure if needed. 7 8 9 10 script name inputs output files output commandLine 11 -------------------------------------------------------------------------------------------------------------------- 12 STGspectra.m coordinate file path STGSpectra.dat (for PHASTA) 13 connectivity file path Matlab savefile for PHASTA spectral data 14 Run Directory 15 number of proccesses 16 17 chooseSTGRand.m PHASTA executable npool optimized STGRand.dat -none- 18 Run directory containter for dmags for each npool 19 Matlab savefile for PHASTA spectral data 20 AnalizeSTG_Rand.m STGRand file Matlab savefile for plotting 2nd moments various additional measures 21 PHASTA execuatble Matlab savefile for plotting 1st moments 22 Run directory 23 Matlab savefile spectra 24 plotBias_TC.m Matlab savefile plotting(2) --none-- plots 25 (Optional) Matlab savefile for TC to bias 26 analyzeTC.m Matlab savefile TC->bias --none-- plots 27 Analyze_Dmag.m STGRand file Dmag for that STGRand 28 Matlab savefile spectra 29 PlotSTG_meanFeilds.m Matlab savefile plotting(1) --none-- 30 31 32 helper functions: 33 ----------------- 34 35 importCords.m 36 importD2Wall.m 37 interpSTG.m 38 investigateMesh.m 39 findPlaneInfo.m 40 getKmax.m 41 getKmin.m 42 importSPECTRApartfile.m 43 importSTGInflow.m 44 importSTGRand.m 45 getSpectra.m
Pre-Proccessing/Preparation of Auxiliary Files
Boundary Condition Attribute
Within the simmodeler or meshing tool, the face that is to have the STG boundary condition must have a specified SurfID attribute that will later match the specified in solver.inp. A typical value that has been given to this boundary condition within our group is 411.
Creation of STGInflow.dat
This auxiliary file will be required not only for the running of PHASTA, but also, will be required for the random number optimization. The user will need mean flow field data at the position of the inflow from some preliminary simulation such as RANS data or data repository. The file format itself can take a few different forms, but the primary is given as the 14 columns version:
N d2wall U_1 U_2 U_3 uu vv ww uv uw vw sclr1 sclr2 eddy_length dissipation ...
Where:
-
Nis the number of rows that this file contains. It is the number of nodes in the specially varying direction. -
d2wallis the distance to the wall -
U_iare the three components of the average velocity field -
uu, vv, ww, uv, uw, vware the Reynolds' stress tensor components -
sclr1andsclr2are the scalars that represent the turbulence model.- This can be k and omega for SST or just nu^tilde for SA
-
eddy_lengthis the length scale of the energy containing eddies -
dissipationis the TKE dissipation term- The latter two are used to form the model spectra used by the STG method
The input profiles are varied by the type of auxiliary simulation. So the post-processing of that preliminary simulation will be varied and so the generation of this data is left to the user. However for creating the STGInflow.dat file it is useful to know the format that the fortran file reader is setup for. The below chunk of MATLAB code is useful once the profiles are known:
nPoints = length(d);
[dSort,index]=sort(d);
fid=fopen('STGINFLOWNAME.dat','w');
fprintf(fid,'%d\n',nPoints);
for j=1:nPoints
i=index(j);
tmp=[d(i) u(i,:) R(i,:) nut(i) lt(i) eps(i)];
if (j~=nPoints)
fprintf(fid,'%.12E %.12E %.12E %.12E %.12E %.12E %.12E %.12E %.12E %.12E %.12E %.12E %.12E\n',tmp);
elseif (j==nPoints)
fprintf(fid,'%.12E %.12E %.12E %.12E %.12E %.12E %.12E %.12E %.12E %.12E %.12E %.12E %.12E',tmp);
end
end
fclose(fid);
Creation of STGSpectra.dat
This file will contain the spectral data at each nodal point within the mesh that is attributed with the STG attribute. To create this file 1) the case's run directory must be prepared and 2) we will use the first of several scripts within the STGInit subdirectory of the github remote repository for PHASTA. Within MATLAB open STGspectra.m. The following is what you should see from the header of the file:
1 %% Create STGspectra.dat
2 % This script will produce spectral data for the implementation in PHASTA
3 % This script's intention is to move the spectral data calculation out of
4 % PHASTA so as to not require a call to PHASTA to gather such data. (A
5 % wasteful thing to do when large meshes are investigated)
6
7 % Authors: Ken Jansen, John Patterson
8 % Version: 1.0
9 % Last mod: 7/3/2020
10
11
12 %STGspectra('exampleInput/LES_DNS_ReL1M_2d_KEJ_SAVEFILE.mat','exampleInput/LES_DNS_ReL1M_2d_KEJ.cnnTet',1,'/projects/tools/Models/STG/STG_tests/Post/analizeRand/RunTetRand_newRAND_2/',8)
13 function []=STGspectra(x_Path,conn_PATH,d2wall_Path,Run_Dir,nprocs)
14 tic
15 %% Preliminary Settings
16 nu=1.5E-5;
17
18 %Specify how the d2wall field may be constructed
19 d2wall_tech='load';%'calculate ana';
20 %Specify (if nessicary), the path of the nodal data of the d2wall field
21 d2wall_Path='d2wall_ana.mat';%'none';
22
23 %Specify the method to determine the nodes of the STGInflow plane
24 STG_node_choose='min_x';
25
26 %Spesify the method by which the mesh data is structured
27 struct_Spe='structured';
28
29 %Settings found in solver.inp that are nessisary for using the
30 %STGInflow
31 deltaBL=0.05;
32 alphaWave=0.01;
33 setEps=17251.0;
34
35
36 %% Unfolding
37 nprocs_Dir =strcat(num2str(nprocs),'-procs_case/');
38 [x,ien,surround_nodes] =importCords(x_Path,conn_PATH);
39 d2wall =importD2Wall(d2wall_tech,x,d2wall_Path);
40
41
42 %% Load in STGInflow and interpolate onto the inflow mesh
43 STGInflow_Path=[Run_Dir,nprocs_Dir,'STGInflow.dat'];
44 STGInflow=importSTGInflow(STGInflow_Path);
45
46 [U,V,W,uu,vv,ww,uv,uw,vw,sclr1,sclr2,lt,eps,ch_decomp]=...
47 interpSTG(STGInflow,x,d2wall);%x(:,2));
48 if(~isnan(setEps))
49 eps=ones(length(x),1).*setEps;
50 end
This will be the standard format for many of the scripts in this subdirectory.
That is,
- Description/ Authors (ln 1->9)
- function call/ function call example (ln 12->13)
- Settings for each run (ln 15->33)
- Loadins from other files (ln 36->47)
- Execution of the script (ln 48->end)
The user-controlled inputs is the content in the function call and the settings below:
function call
There are five inputs to this function call
- x_Path (str)
- conn_Path (str)
- x_Path and conn_Path are strings that point to files that are able to be interpreted by importCords.m. When these files read-in a .crd .cnn type files, they will output not only what is required for the STGspectra function call, but also a MATLAB .mat file that will append the names of the original data with '_SAVEFILE'. Loading in this .mat file is MUCH quicker than the .crd and .cnn raw data.
- d2wall_Path (str)
- d2wall_Path is a path to a MATLAB datafile containing the distance to the wall for each node in the mesh Unlike x_Path and conn_Path these two files are not required for the script to run properly SEE Settings for each run BELOW
- nprocs (int)
- this will be the number of processors that the case will use in the execution of PHASTA. In this script it is used to import the STGInflow.dat file.
Settings
There are 8 inputs to this function call
- nu: the kinematic viscosity of the fluid
- d2wall_tech: This is the technique by which the code will make the distance to the wall field. There are currently 5 options to this:
- x_dir: Takes the x coordinate as the distance to the wall
- y_dir: Takes the y coordinate as the distance to the wall
- z_dir: Takes the z coordinate as the distance to the wall
- calculate_ana: This will calculate the distance to the wall via the function handle that is specified in importD2Wall ln 17 (Please modify this if you wish to create your own d2wall function) Like the importCoords file this will output a matlab file containing the d2wall at d2wall_ana
- load: this will load in the path specified from d2wall_Path
- d2wall_Path: If you choose to load in a specific d2wall field that has been already saved this will be the path to that file
- struct_Spe: Currently only 'structured' is supported. This is the orientation of the mesh.
- deltaBL: This is the boundary layer thickness at the STG boundary condition.
- alphaWave: This is the logarithmic growth of the spacing between one wavenumber and the next. It may be set from 0.05 to 0.01. Normally we set this to 0.01.
- setEps: This is set to the dissipation rate of the desired inflow spectrum.
Creation of STGRand.dat
Creating STGRand.dat is done within the chooseSTGRand.m MATLAB script. This script is meant to be run at the particular case's run directory (ie. above the n-procs_case dir). This script contains eight sections:
- Choose STGRands
- Parameter set
- Set up random sets and collect spectra data
- Collect Target STG profiles
- Set a custom spectra
- interpolate the target cholesky decomposition to the mesh grid in geombc
- Begin the choice of random number sets
- Print STGRand.dat
Of these sections, the layman user should be concerned with the first two.
1 %% Choose STGRands
2 % This script chooses Random varaibles For STGRand.dat. The output will be
3 % npool number of optimized random number sets, saved in STGRand files, As well as
4 % a display of the bias accourding to developed error measures for bias.
5 % Each set is measured against a specified spectra, and target covaraince.
6
7 % Authors: Ken Jansen, John Patterson
8 % Version: 1.0
9 % Last mod: 6/30/2020
10 % Order Spec: Sigma first, intellegent design d, intellegent design
11 % low-to-high wavemode
12 tic
13 clear all
14 close all
15 delete(gcp('nocreate'))
16 %% Parameter set
17
18 %These set the number of proccesses will search for optimal random number
19 %sets (npool) and the number of random number sets each procces will
20 %search through (nRand). NOTE:
21 %1) viz002 and viz003 have 8 and 10 proccesses respectivally for matlab.
22 %2) nRand must be greater than or equal to the nkwave that PHASTA
23 %produces from getSpectra.m
24 npool=12;
25 nRand=100000;
26
27 %This choice will allow the user to specify a costom spectra, IF we want
28 %the spectra from PHASTA set this to 'none'
29 specChoice='none'%max3';%'mid8%';
30
31 % nprocs: # in #-procs directory the spectral data is to be
32 % pulled from. If you want to recreate the spectral content via PHASTA, it
33 % is reccommended that the user chooses the largest partition they have
34 % availible
35 % The spectraPATH parameter sets one of a few things:
36 %---------USE 1 for this script if the version of PHASTA lacks a readin from STGSpectra.dat----------------------------------------
37 % 1) If the user has no prior knowledge of the spectra created from
38 % PHASTA then make this parameter the path to the run directory with
39 % RECREATE appended to the end.
40 %---------USE 2 for this script if the version of PHASTA has a readin from STGSpectra.dat----------------------------------------
41 % 2) If the user has Spectral output from PHASTA (IE if (1) was choosen
42 % and you wish to reuse the data) make this path the run dir OR if the
43 % spectral output from STGspectra.m has been created for use in
44 % PHASTA.
45 nprocs=8;
46 spectraPATH='/projects/tools/Models/BoeingBump/DNS/FlatPlate/Meshing/matchedReader_CRS_4delta6-15/Chef/8-8-Chef/RunPHASTA/';
47 run_Dir=strsplit(spectraPATH,'RECREATE');run_Dir=cell2mat(run_Dir(1));
48
49 % These weights will specify which target covarience quantities are
50 % most-highly valued
51 %Wstress: The applied penelty for a particular component of the reynolds stress that is
52 % off
53 %nlocs: The node number set in the spacial variation dir. that we choose to be the
54 % control points (The points where we get the covariance correct)
55 %weights: The applied penelty for a particular component of the reynolds stress that is
56 % off, at the positions y(nlocs)
57 Wstress=[1,25,1;25,5,1;1,1,1]; % start with equal on all
58 nlocs= [44,57,72,75,85,105];
59 weights=[1, 2, 2, 2, 2, 1];
60 weights=weights./sum(weights) ;
61
62 %physical quantites for normalized units
63 rho=1;
64 mu=1.2502E-5;
65 nu=mu/rho;
66
67 % Specify the output STGRand files to be made
68 STGRand_basename='STGRand_REDO_6_12_hex_JP_nRand100k';
Choose STGRands
This section goes over the description of the script: This script chooses Random variables For STGRand.dat. The output will be npool number of optimized random number sets, saved in STGRand files, As well as a display of the bias according to developed error measures for bias. Each set is measured against a specified spectra, and target covariance.
Parameter set
This section of the code are the primary variables to set when optimizing a particular random number set for a particular case. The script has descriptions of each of the variables but this documentation will attempt to provide more detail with regard to each one. A few variables that pertain to the particular case you wish to optimize to:
- specChoice: This string will determine which spectra you wish to optimize to, the entire set of choices are the cases in Set a custom spectra section of the code
- 'none' is the most common choice for this option because the user is not commonly using their own spectra that hasn't been calculated in the proceeding getSpectra.m function call.
- 'max3' is using the the maximal intensity wavenumbers as their spectra to optimize to.
- 'mid8%' is using the middle region of the spectra to optimize to
- nprocs: This integer is the number of processes that the case will have. This is mostly used for read-in scripts of the STGSpectra.dat and STGInflow.dat files
- spectraPATH: This string is the path to where the spectra MATLAB file has been created from the previous step, or the PHASTA run that prints out the spectra file 'spectra00n'.
- run_Dir: This string is the path to the run directory containing the n-procs case that you wish to investigate.
- STGRand_basename: This string will be the name that will specify the npool random number output filenames.
- rho: This is the physical density of the fluid.
- mu: This is the physical viscosity of the fluid.
- u0: this double is the free-stream velocity of the fluid
In addition there are a few parameters that pertain to 1) what the optimization process is optimizes to, 2) the amount of searching to de, and 3) what computational resources the user wishes to give to MATLAB to perform the search. Some choices are better than others. In the order in the script:
- npool: This integer is the number of processes that the user wishes to allow MATLAB to use to perform the search. The maximum that the user can ask for is 8 on viz002 and 12 on viz003.
- nRand: This integer is the amount of random number sets that each of the MATLAB proccesses will choose from. These random number sets only pertain to the sigma vector. The d-vector is a unique set based off of the best set of random sigmas.
- Wstress: This 3x3 matrix of doubles instructs the optimization to which stress components to preferable to. A choice of [1,25,1;25,5,1;1,1,1]; is good for a flat plate case where the stream-wise direction is aligned with x_1 and span-wise direction is aligned with x_3. This is the case because 1) the <u_1u_2> component of the Reynold's stress contributes much more to PHASTAs ability to recover downstream, and 2) the diagonal components in the tensor are the easiest to get correct so often a sufficient <u_1u_2> implies sufficient diagonal components .
- nlocs: This array of integers are the node numbers from the wall that the method chooses to optimize to. The preferable nodes to optimize to are the nodes that approximate the peak of the Target Reynolds stress components. The length of this array can be up to the number of nodes in the y-direction.
- weights: This array of integers is how much importance each of these nlocs get when optimizing. The length of this array is the length of the nlocs array.
Output and what to lookout for
The result of this file will output npool number of valid STGRand.dat number sets named according to the STG_basename and the process that created it. Additionally, the method will output in the command window the Dmag for each process. The lowest of these numbers is the best STGRand.dat file that was created from the MATLAB proccesses.
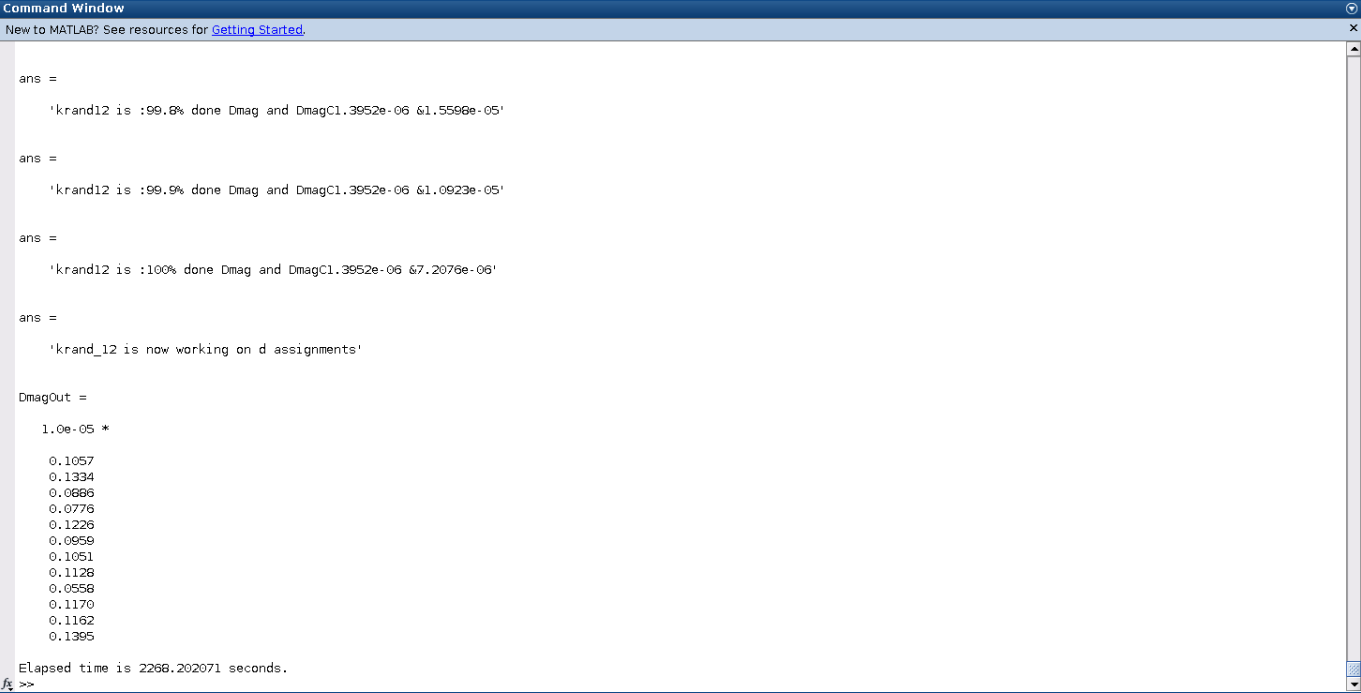
Pre-Simulation-Analyzation
This section is meant to outline usage of the tools that are available to analyze the random number sets that are chosen (whether they be optimized or not).
analyzeSTG_Rand.m
The first and most important of these tools is analyzeSTG_Rand.m, the preceding analyzation scripts, (with the exception of Analyze_Dmag.m) may not run without it. Primarily, this script will create a time-spamwise average of first and second order statistical moments of the full field stochastic velocity field. Like chooseSTGRand.m, this analization will take some time (although MUCH less time than an actual simulation), and therefore will use multiple processes to collect the statistical data. It is advisable to start with a smaller resolution (IE smaller nz and larger nsk) in order to make sure that your inputs are working properly before making a full simulation, as these nominally take on the order of 1-2 days to complete with the highest resolution.
Many of the knobs to turn in this section are the same as in chooseSTGRand.m and getSpectra.m. All of the controls that the user should be concerned with are those in the Parameter set section:
- nprocs: This integer is the number of processes that the case will have. This is mostly used for read-in scripts of the STGSpectra.dat and STGInflow.dat files
- spectraPATH: This string is the path to where the spectra MATLAB file has been created from the previous step, or the PHASTA run that prints out the spectra file 'spectra00n'.
- run_Dir: This string is the path to the run directory containing the n-procs case that you wish to investigate.* specChoice: This string will determine which spectra you wish to optimize to, the entire set of choices are the cases in Set a custom spectra section of the code
- 'none' is the most common choice for this option because the user is not commonly using their own spectra that hasn't been calculated in the proceeding getSpectra.m function call.
- 'max3' is using the the maximal intensity wavenumbers as their spectra to optimize to.
- 'mid8%' is using the middle region of the spectra to optimize to
- STGInflow_filepath:
- dt: This is the timestep that the PHASTA simulation will see.
- rho: This is the physical density of the fluid.
- mu: This is the physical viscosity of the fluid.
- u0: this double is the free-stream velocity of the fluid
- wavegrowth: This is the growth rate of the custom spectra, usually this is 0.01 to 0.05.
- bl: This is the boundary layer height on the STG boundary conditon
- x: This is the x_1 coordinate of the STGInflow plane.
- zmin: This is the minimum z coordinate of the STGInflow plane.
- zmax: This is the maximum z coordinate of the STGInflow plane.
- nz: This will be the number of z coordinates that the simulation will average over. This number must be divisors of the number of nodal points of the mesh in the z direction.
- nsk: This is the number of y points that the averaging will skip in the y-direction. The minimum this can be is 1.
- tstep: This is the number of timesteps that are required for a flow through.
- tau: This array will contain the flow-through times that the user desires to be collected
- weights: This array of integers is how much importance each of these nlocs get when optimizing. The length of this array is the length of the nlocs array.
- nloc1->nlocn: This array of integers are the node numbers from the wall that the method chooses to optimize to. The preferable nodes to optimize to are the nodes that approximate the peak of the Target Reynolds stress components. The length of this array can be up to the number of nodes in the y-direction.
- momANA: This integer array should contain 1 if the mean flowfield statistical data is desired. If the 2nd moment data is desired, this integer array should contain 2.
The only input to the function call is the case which determines the following:
- path: This string is the path the STGRand.dat file that the user wishes to analyze.
- saveName1: This string will be the name given to the .mat file that contains the 1st order statistical moments of the simulation.
- saveName2: This string will be the name given to the .mat file that contains the 2nd order statistical moments of the simulation
The function call looks as follows:
1 %% Function: analyzeSTG_Rand
2 % This funtion is meant to analyize a particular random number set and
3 % output that data into a save file so that the plotting scripts may plot quickly
4
5 % Authors: Ken Jansen, John Patterson
6 % Version: 1.0
7 % Last mod: 9/10/2020
8 function []=analyzeSTG_Rand(caseN)
9 delete(gcp('nocreate'))
10 tic
11 %% Parameter set
12 %Every thing in this section of the code must be filled out in order for
13 %the script to provide meaningful data. The case choice looks more daunting
14 %than it is, specify a STGRand.dat to readin. The cases where used so that
15 %we could assess multiple cases on different runs of the function at the
16 %same time without modification of the code.
17
18 % nprocs: # in #-procs directory the spectral data is to be
19 % pulled from. If you want to recreate the spectral content via PHASTA, it
20 % is reccommended that the user chooses the largest partition they have
21 % availible
22 % The spectraPATH parameter sets one of a few things:
23 % 1) If the user has no prior knowledge of the spectra created from
24 % PHASTA then make this parameter the path to the run directory with
25 % RECREATE appended to the end.
26 %---------USE 2 for this script----------------------------------------
27 % 2) If the user has Spectral output from PHASTA (IE if (1) was choosen
28 % and you wish to reuse the data) make this path the run dir
29 nprocs=8;
30 spectraPATH='/projects/tools/Models/BoeingBump/DNS/FlatPlate/Meshing/matchedReader_CRS_4delta6-15/Chef/8-8-Chef/RunPHASTA/';
31 run_Dir=strsplit(spectraPATH);run_Dir=cell2mat(run_Dir(1));
32
33 %This choice will allow the user to specify a costom spectra, IF we want
34 %the spectra from PHASTA set this to 'none'
35 specChoice='none';
37 %Parameters from Solver.inp and dirived from STGInflow.dat
39 STGInflow_filepath='../CRS_d4-Optimal/STGInflow.dat';
40 u_Tau=0.046987317422423;
41 u0=1;
42 dt=7.5E-4;%1.5E-6;
43 rho=1;
44 mu=1.2502E-5;
45 nu=mu/rho;
46 distPLUS=u_Tau/nu;
47 velPLUS=1/u_Tau;
48 wavegrowth=0.01;
49 bl=0.113;
50 lemax=2*1.53415*bl;%SAVE THIS
51 DOM_LENGTH=1.5;
52 flowthroughDIST=DOM_LENGTH*(3/2);
54 %%Parameters specific to the fidelity of the analysis desired
56 % not used but never forget kemin=2*kmin;
57 %streamwise component of the STGInflow
58 x=-3.1;
59 %spanwise mesh of the STGInflow
60 zmin = 0; zmax = 0.34;Lz = zmax-zmin; nz=213; %85 gives every 4th pt; to get all use -> 340;
61 % to avoid using the periodic point (end) twice requires that we either
62 % start shifted dx or end dx before zmax
63 dzReal=Lz/nz;
64 dzS=nz/213*dzReal;
65 zpos=0:dzS:(zmax-dzS);
66 % nsk: Number of skipped nodes in y before assesing the next line of spanwise
67 % coords
68 nsk=2;
69 %dt: simulation time step
70 %tstep: timestep for gathered time convergance data
71 dt=7.5E-4;tstep=4500*dt;
73 %tau: physical times by which we want to time average to
74 tau=tstep.*[0.25,1,2,4];%,8,16,32,64,128,256,512,1024,2048,4096,8192];
76 %debug flag
77 debug=0;
79 %These are parameters set for bias estimates
80 ystart=30;
81 nyskip=5;
82 nycalc=10;
83 %The number of noders to order the worst perpetrators
84 nmost=10;
85 %weights as it is in chooseSTGRand.m
86 weights=[1,2,2,1];
87 %
88 nloc1=28;nloc2=40;nloc3=49;nloc4=59;
89 %Desired moments to be analized.
90 % ex: [1] will produce only 1st order moments
91 % ex: [1 2] will produce 1st and 2nd moment data
92 momANA=[1,2];
93
94
95 %%Load in Random number set
96 switch caseN
05 case 19
06 path='STGRand_REDO_6_12_hex_JP_nRand100k9_26-Feb-2021.dat';
07 saveName1='STGRand_REDO_6_12_hex_JP_nRand100k9_26-Feb-2021_meanSave'
08 saveName2='STGRand_REDO_6_12_hex_JP_nRand100k9_26-Feb-2021_varSave';
09 STGRand=importSTGRand(path);
10 end
11 nkwave=STGRand(1,1);
12 d =STGRand(2:end,1:3);
13 phi1=STGRand(2:end,4);
14 sig=STGRand(2:end,5:7);
15
16 %% load in the spectra
plotBias_TC.m
This script output plots that are meant to demonstrate bias and the expected time convergence relative to the second order stresses. Better visualization of the time convergence can be visualized via analyzeTC.m. That script requires output from this script. In a matlab file save the SpanWise_rms4k8k and SpanWise_rms4k8kMax with the loadedCase string appending it. Do the same in the same matlab file with other cases you wish to compare against.
After this script has executed enter the following in the command line to save the required data for analyzeTC.m:
save(strcat(loadedCase,'_Spanwise_rms'),'SpanWise_rms4k8k','SpanWise_rms4k8kMax').
As this script is mostly to plot desired quantities, the code under ln 184 is for formatting the plots for display. The lines below 85 are to create the root mean squared data that is useful for the analyzeTC.m script, but is no use for the users to know about. The user need only specify (3) inputs:
- caseN: This will specify the MATLAB file that was the output from analyzeSTG_Rand.m you wish to plot.
- plotGoals: 1 will specify if the target statistical profiles are to be included in the plots.
analyzeTC.m
This script, like plotBias_TC.m is mostly a plotting script. The input needs to be the MATLAB file that is created from the save command specified in the plotBias_TC.m section.
Analyze_Dmag.m
This script is meant to re-output the Dmag error metric used to determine the best STGRand.dat of the npool STGRand.dat s that where output from chooseSTGRand.dat. It is useful if the user creates a number of STGRand.dat files and forgets which one is the best of the group. The inputs to this file therefore mimics the inputs of the chooseSTGRand.m script.
PlotSTG_meanFeilds.m
This file plots the first order statistical moments that are the output of analyzeSTG_Rand.m MATLAB script. The inputs here will therefore be exactly the same as that of the analyzeSTG_Rand.m MATLAB script.
Running the code
In addition to the case set-up the additional auxiliary files within the n-procs case directory that are needed for PHASTA to use the STG boundary condition are STGInflow.dat, STGRand.dat, and STGSpectra.dat. The only other specifications needed are within the Solver.inp file.
Solver.inp
This is the final remaining step in the workflow to include the STG inflow boundary condition. Below are the default conditions pertaining to the STG boundary and initial condition
137 BOUNDARY AND INITIAL CONDITIONS
138 {
146 Ramp Down Nut in STG : False
147 x Position Nut STG Ramp 0 : 0.123
148 x Position Nut STG Ramp 1 : 0.456
151 Load and set STG IC: False
153 }
547 #STG input Parameters
548 {
549
550 Use STGinflow BCs: False
551 STG Dump Spectra:0
552 STG Start Step: 0
553 STG Turn Off Fluctuations: False
554 STG SurfID: NODEFAULT
555 STG BL Height: 1.0
556 STG U_0: 1.0
557 STG Number of Modes: 1.0
558 STG Mode Growth: 1.0
559 STG Mesh Growth: 1.0
560 STG Dissip. Rate: 1.0e6
561 STG Channel: False
562 STG SST Method: 1
563 STG SST uTau: 1.0
564 STG SST Eddy Viscosity Factor: 1.0
565 Collect STG Energy Spectrum Data: False
566 STG Energy Distance Tolerance: 1.0
567 Energy Spectrum at Distances: 0.0 0.0 0.0
568 Number of Points for Eng. Calculation: 0.0
569 }
Many of these are known from the preceding steps with the exception of a few:
- STG Dump Spectra: When this is 1 a PHASTA run will dump spectra files that getSTGspectra.m can read-in.
- STG Start Step: this is the timestep that PHASTA will turn on the STG boundary condition
- STG Turn Off Fluctuations: Turning this to True will provide the mean flowfield at the inflow without the STG fluctuations.
- STG SurfID: This will be the SurfId that was assigned at the problem definition step of the workflow.